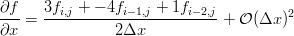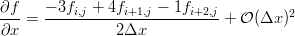Background and Motivation
The problem I am interested in: leverage high-level symbol manipulation capabilities in a computer algebra system (CAS) to generate compile-able code for number crunching. More specifically, I am interested in generating routines for numerical partial differential equation (PDE) solvers. This is not a new idea, significant work was accomplished starting in the early 1980s [1, 2, 3], continued into the late ’80s [4, 5, 6], and early ’90s [7]. However, those capabilities have not been consolidated and formally incorporated into my open-source CAS of choice, Maxima (though there is fairly recent work implementing similar ideas for Mathematica [8, 9]). The early work [1] quotes Hamming’s Turing Award speech [10] for motivation:
We seem not to be able to use the machine, which we all believe is a very powerful tool for manipulating and transforming information, to do our own tasks in this very field. We have compilers, assemblers, monitors, etc. for others, and yet when I examine what the typical software person does, I am often appalled at how little he uses the machine in his own work.
I had been thinking about this problem for a while because of my interest in method of manufactured solutions (MMS) for code verification. I thought things were really going to take off when I came across Cook’s 1990 technical report describing code generation for PDE solvers using A Livermore Physics Applications Language (ALPAL), which was built on top of DOE-MACSYSMA. ALPAL aimed to be a general purpose domain specific language for turning symbolic descriptions of PDEs into code. Here’s a solution to the problem already developed! Of course, it couldn’t be that easy. I asked the Maxima mailing list if anyone knew what became of this effort: ALPAL question to Maxima list, and ended up answering my own question by getting a hold of some of the old ALPAL devs: ALPAL answer to the list. Unfortunately, there is
a long history behind the divergence between different versions of MACSYMA (and different groups of developers) that mitigates against this old code ever working with the fork that became Maxima (should an archive of the source ever actually turn up, update: it did, see comment)
[11, 12, 13].
As you may have guessed from this post’s title, and the difficulties described in the previous paragraph, I’ve decided to pursue a more toolbox approach rather than (re)implementing a domain specific language and associated decision logic. The working title for my toolbox of utilities will be NALPAL, both as a nod to the valuable historical work in this area, and an indication of the path not taken.
Wirth categorizes three main approaches to systems for numerical PDE solution: black-box systems, subroutine packages, and code generation aids [1]. Black-box systems are intended for the novice user, and tend to be constrained to very specific problem types or incorporate a great deal of decision logic. Subroutine packages provide reusable building blocks, but require more knowledge and coding on the part of the user, and tend to remain problem specific. Code generation aids require the user to be an expert in numerical analysis, and tend not to automate any of the analytical work that precedes the code generation step.
Wirth’s suggested approach is a Unix-like toolbox approach where standard subroutine libraries are used when it makes sense to do so, and utilities for doing custom code generation are written in the CAS environment. The alternative is to create a full language, and incorporate the semantics and decision logic to automate the entire process for the novice user. The work of Wirth, Steinberg and Roache is a good example of the former approach, and Cook’s work on ALPAL is an example of the latter (though his early work [3] is more along the lines of the toolbox approach, going so far as to say “the viewpoint taken is that one should be able to start with integro-differential equations and end up with optimal FORTRAN code segments. The key in achieving this goal was to tackle only the specific algebraic problem-i at hand, and not to attempt to provide tools to cover every conceivable numerical scheme.”). The toolbox approach keeps more of the work for the problem solver in set-up/tear-down/customization, while the language approach loads more of the burden on to the domain specific language developer. Wirth’s second objective sums up well the approach that I think makes the most sense (emphasis original):
Build productivity enhancing tools of broad applicability for the expert user so that efficient, special purpose PDE codes can be built reliably and quickly, rather than attempt to second guess the expert and build general purpose PDE codes (black box systems) of doubtful efficiency and reliability.
There are still useful things to learn from ALPAL even if we have chosen the less ambitious road, since the problem being solved is the same. A basic description of the ALPAL use-case
[7]:
- Take as input a PDE description, along with boundary and initial condition definitions
- Discretize the PDE
- Analyze the result (e.g. for stability)
- Calculate the Jacobian (needed for Newton methods in implicit time-integration or non-linear boundary value problem (BVP)s)
- Generate code
Even if we don’t have a full-featured language, the user of our set of utilities will still be interested in accomplishing largely these same steps. In fact, Wirth’s early work gives these steps (reproduced here verbatim) [1]:
- Manipulate the set of partial differential equations to cast them into a form that is amenable to numerical solution. For vector PDEs, this might include vector differential calculus operations and reexpression in scalar (component) form, and the application of a linearization approximation for non-linear PDEs.
- Discretize the time and space domain, and transform the partial differential operators in the PDEs into finite difference operators. This transforms the partial differential equations into a set of algebraic equations. A multitude of possible transformations for the differential operators are possible and the boundary conditions for the PDEs also must be appropriately handled. The resulting difference equations must be analyzed to see if they form an accurate and numerically stable approximation of the original equation set. For real world problems, this analysis is usually difficult and often intractable.
- After choosing a solution algorithm from numerical linear algebra, the finite difference equations and boundary conditions are coded in a programing language such as FORTRAN.
- The numerical algorithm is then integrated with code for file manipulations, operating system interactions, graphics output, etc. forming a complete computer program.
- The production program is then executed, and its output is analyzed, either in the form of numerical listings or computer-generated graphics.
Wirth goes on to say (emphasis added),
With continuing advances in computer technology, the last step in this process has become easier. For a given class of problems, answers can be calculated more quickly and economically. More importantly, harder problems which require more computational resources can be solved. But the first four steps have not yet benefited from advances in computer performance; in fact, they are aggravated by it.
Also, this additional bit of motivation from Chapter 5,
Taken together with the software described in other chapters, these tools allow the user to quickly generate a FORTRAN code, run numerical experiments, and discard the code without remorse if the numerical results are unsatisfactory.
This is similar in sentiment to the idea of numerical throw away code.
PDE Code Gen Recipes
With the problem background and motivation set, the rest of this post will focus on pulling out useful Maxima recipes demonstrated by folks who have generated FORTRAN from MACSYMA with intent to inflict grievous arithmurgical damage on hapless PDEs.
Much work is done in most of these articles from the 1980s to avoid array references and break up expressions by hand to reduce the computational cost in MACSYMA. Also, MACSYMA’s knowledge of the chain rule is not used for calculating the transformations because of an explosion in the number of terms [5], rather an identity for the derivative of a matrix inverse is used. A lot of this effort seems unnecessary today because speed and memory have improved so much. However, the basic approach and variables used to define the problem is still relevant (compare with this example of Burgers’ equation on a curvilinear grid):
dep : [f, sigma]; /* the dependent variables */
curvi : [xi, eta, zeta]; /* the curvilinear coordinates */
indep : [x, y, z]; /* the independent variables */
depends(curvi, indep);
depends(dep, curvi);
nn : length(indep);
eqn : sum(diff(sigma * diff(f, indep[i]), indep[i]), i, 1, nn);
curvi : [xi, eta, zeta]; /* the curvilinear coordinates */
indep : [x, y, z]; /* the independent variables */
depends(curvi, indep);
depends(dep, curvi);
nn : length(indep);
eqn : sum(diff(sigma * diff(f, indep[i]), indep[i]), i, 1, nn);
The result is rather large, but it illustrates Maxima’s knowledge of the chain rule. Of course, generally it is easiest to compute  rather than
rather than  for an arbitrary grid, so you need to make substitutions based on the inverse of the Jacobian of transformation. In Maxima we might do something like
for an arbitrary grid, so you need to make substitutions based on the inverse of the Jacobian of transformation. In Maxima we might do something like
 rather than
rather than  for an arbitrary grid, so you need to make substitutions based on the inverse of the Jacobian of transformation. In Maxima we might do something like
for an arbitrary grid, so you need to make substitutions based on the inverse of the Jacobian of transformation. In Maxima we might do something like
J : zeromatrix(3,3);
for j : 1 thru 3 do (
for i : 1 thru 3 do (
J[i,j] : ’diff(indep[j],curvi[i])
)
);
K : zeromatrix(3,3);
for j : 1 thru 3 do (
for i : 1 thru 3 do (
K[i,j] : diff(curvi[j], indep[i])
)
);
grid_trans_subs : matrixmap(”=”, K, invert(J));
/* making substitutions from a list is easier than from a matrix */
grid_trans_sublis : flatten(makelist(grid_trans_subs[i],i,1,3));
for j : 1 thru 3 do (
for i : 1 thru 3 do (
J[i,j] : ’diff(indep[j],curvi[i])
)
);
K : zeromatrix(3,3);
for j : 1 thru 3 do (
for i : 1 thru 3 do (
K[i,j] : diff(curvi[j], indep[i])
)
);
grid_trans_subs : matrixmap(”=”, K, invert(J));
/* making substitutions from a list is easier than from a matrix */
grid_trans_sublis : flatten(makelist(grid_trans_subs[i],i,1,3));
which gives us a list of nine equations we can use to make substitutions so that all our derivatives are with respect to the computational coordinates.
trans_eqn : subst(grid_trans_sublis, eqn) $
/* Evaluation took 0.0510 seconds (0.0553 elapsed) using 265.164 KB. */
trans_eqn_factor : factor(trans_eqn) $
/* Evaluation took 2.4486 seconds (2.5040 elapsed) using 48.777 MB. */
/* Evaluation took 0.0510 seconds (0.0553 elapsed) using 265.164 KB. */
trans_eqn_factor : factor(trans_eqn) $
/* Evaluation took 2.4486 seconds (2.5040 elapsed) using 48.777 MB. */
Factoring the result starts getting on up there in compute time and memory, but still not untractable, or even that uncomfortable for an interactive session. Of course we still haven’t made any difference expression substitutions, and that will expand the number of terms even further. The assumption that the grid is arbitrary is a decision point that would have to be dealt with in a black-box style implementation. Best to leave it to the (hopefully) knowledgeable user.
As an aside, linearization is another example of an assumption that is probably best left to the user. Wirth assumes that a linearization is required to solve nonlinear PDEs, whereas Cook provides for calculating the system Jacobians needed for Newton methods (of course Wirth does note that this is just one approach that could be used, and the tools are flexible enough to be extended to other methods). Using the Jacobian is a more modern approach made practical by the successful development of Newton-Krylov methods. It is impossible to predict the future development of numerical methods that will change the choices that need to be made in discretizing PDEs (though that doesn’t prevent speculation [14]), so a flexible toolbox approach is again indicated.
Once a PDE is defined substitutions of finite differences for partial derivatives must be made to create the discrete approximation. Wirth uses a function called DISCRETIZE which uses the dependencies list (created by a call to depends) to substitute indexed variables for the independent variables. Then the partial derivatives of the indexed variables are replaced by finite difference operators. The substitutions of difference expressions is controlled by using Maxima’s pattern matching rules. The basic recipe is
- Define dependencies between independent and dependent (and possibly computational coordinates)
- Associate a list if indices with the coordinates
- Define rules that transform differential terms into difference terms with the appropriate index shifts and constant multipliers corresponding to the coordinate which the derivative is with respect to and the selected finite difference expression
- Apply the rules to the PDE to give a finite difference equation (FDE)
- Use Maxima’s simplification and factoring capabilities to simplify the FDE
- Output the FDE in FORTRAN format and wrap with subroutine boilerplate using text processing macros
The boilerplate is a set of standard templates for a particular solution procedure. The example Wirth uses is an alternating direction implicit (ADI) scheme. A more modern example might be a Newton-Krylov scheme. Wirth describes an environment, Fast FORTRAN Programing (FFP), whose goal is to move computational physics out of a “cottage industry” state. He describes it thusly, “The system consists of two major components: a subroutine library and a command language for building, compiling and running codes.” Based on my experience, that sounds a whole lot like Scipy, which is built on top of Python, and packages together many of the classic scientific computing libraries.
Pattern matching in Maxima is relatively straight-forward. For instance, say I have a function that I’d like to use to calculate my derivatives (such as one based on the discrete cosine transform (DCT)), I could declare patterns that replaced derivatives with function calls.
matchdeclare([fmatch,xmatch,nmatch],all),
defrule(deriv_subst_1, ’diff(fmatch,xmatch,1), diff_1(fmatch,xmatch)),
defrule(deriv_subst_2, ’diff(fmatch,xmatch,2), diff_2(fmatch,xmatch)),
mat_expr : apply1(mat_expr, deriv_subst_1),
mat_expr : apply1(mat_expr, deriv_subst_2),
defrule(deriv_subst_1, ’diff(fmatch,xmatch,1), diff_1(fmatch,xmatch)),
defrule(deriv_subst_2, ’diff(fmatch,xmatch,2), diff_2(fmatch,xmatch)),
mat_expr : apply1(mat_expr, deriv_subst_1),
mat_expr : apply1(mat_expr, deriv_subst_2),
This would result in all of the first and second derivatives in mat_expr being replaced by function calls diff_1 and diff_2 respectively.
The initial set-up steps Cook uses in [3] are roughly the same as those used by Wirth, with the addition of a constant list. The focus is more on what to do with the discrete expressions once they are generated. The basic recipe is
- reduce_cnst()
- gcfac()
- optimize()
- fortran()
The constant list allows reduce_cnst to pull constants out of loops, and optimize pulls out common sub-expressions to speed things up. optimize returns a Maxima block, which is similar to a Fortran subroutine. In fact, turning blocks into subroutines in other languages is a common problem which has been addressed on the Maxima mailing list (e.g.
block2c, see also cform), and as Dan Stanger points out, this is the purpose of the GENTRAN package. GENTRAN is not yet successfully ported to work with Maxima [13] (though there is a gentran directory that ships in the Maxima source tarball if you want to take a look).
The only reason Cook went to the Lisp level in [3] was to reduce the cost of the factor routines (which is less of a concern now), and to make optimize work with lists of expressions (which it does now). One of the examples cites a need for an outrageous 500 million words of computer memory for one calculation, but that’s roughly half what’s in my old desktop computer I got used about a year ago (for about $100). The computing power available to the average amateur has come a long way since 1982.
Both Wirth and Cook assume that an arbitrary orthogonal coordinate system (like spherical, polar, cylindrical, Cartesian, etc.) will be defined. A slightly more modern approach is presented by Roache et al. [4, 5, 6]. They assume an arbitrary curvilinear coordinate system, which may not have analytical scale factors and metrics (i.e. they include the numerical grid generation task as part of the problem statement / solution procedure).
The approach demonstrated in [5] focuses on transforming a set of PDEs to arbitrary curvilinear coordinates described by Laplace’s equation. This approach couples the PDE solution and grid generation methods together. Modern approaches in the engineering world generally assume that the grid will be generated by a separate tool (the fully coupled approach can still be useful for adaptive or moving grids), though solving problems on a single, canonical grid seems to still be common in the sciences. The MACSYMA routines presented there are
- change()
- change variables, convert arbitrary second order differential equation in nn variables to an arbitrary coordinate frame in the variables xi[i]
- notate
- atomic notation for derivatives
- notation(exp,vari)
- primitive atomic notation
- scheme()
- introduce differences of unknowns
- difference(u,f,exp)
- primitive differences, scheme and difference collect the coefficients of the differences and calculate the stencil of the solver and coordinate transformations
- myFORTRAN()
- write the FORTRAN code
A lot of this extra effort is avoidable now, because it is tractable to use Maxima’s knowledge of the chain rule, and the built-in indexed variables and pattern matching facilities.
Conclusion
After digging in to the old literature on generating code from MACSYMA, and trying out a few things with current (v5.20.1) Maxima, it seems like all the pieces you need to generate PDE solving code already ship with Maxima (not touched on in this post were the vector calculus and tensor capabilities that Maxima has as well). I kind of already knew this since I'd been generating solver code in an ad hoc sort of way already. Perhaps there is a place for building up a pattern matching rules database, and maybe boilerplate templates. That seems to be the area that the modern efforts are focused on (e.g. Scinapse claims that it’s rules encoding PDE solution knowledge make up half it’s 120kloc and is the fastest growing part of the code-base [9]). The recipes presented here seem more like a documentation, or tutorial item rather than a possible new Maxima package.
References
[1] Wirth, M. C., On the Automation of Computational Physics, Ph.D. thesis, University of California, Davis, 1980.
[2] Wirth, M. C., “Automatic generation of finite difference equations and fourier stability analyses,” SYMSAC ’81: Proceedings of the fourth ACM symposium on
Symbolic and algebraic computation, ACM, New York, NY, USA, 1981, pp. 73–78.
[3] Cook, G. O., Development of a Magnetohydrodynamic Code for Axisymmetric,
High-β Plasmas with Complex Magnetic Fields, Ph.D. thesis, Brigham Young University, December 1982.
[4] Florence, M., Steinberg, S., and Roache, P., “Generating subroutine codes with MACSYMA,” Mathematical and Computer Modelling, Vol. 11, 1988, pp. 1107 – 1111.
[5] Steinberg, S. and Roache, P. J., “Symbolic manipulation and computational fluid dynamics,” Journal of Computational Physics, Vol. 57, No. 2, 1985, pp. 251 – 284.
[6] Steinber, S. and Roache, P., “Using VAXIMA to Write FORTRAN Code,”
Applications of Computer Algebra, edited by R. Pavelle, Kulwer Academic Publishers, August 1984, pp. 74–94.
[7] Cook, G. O., “Construction of large-scale simulation codes using ALPAL (A Livermore Physics Applications Language),” Technical Report UCRL-102469, Lawrence Livermore National Labs, October 1990.
[8] Husa, S., Hinder, I., and Lechner, C., “Kranc: a Mathematica package to generate numerical codes for tensorial evolution equations,” Computer Physics Communications, Vol. 174, No. 12, 2006, pp. 983 – 1004.
[9] Akers, R. L., Kant, E., Randall, C. J., Steinberg, S., and Young, R. L., Enabling
Technologies for Computational Science, Vol. 548 of The Springer International Series in
Engineering and Computer Science, chap. SCINAPSE: A problem solving environment for partial differential equations, Springer, 2000, pp. 109–122.
[10] Hamming, R. W., “One Man’s View of Computer Science,” Journal of the
Association for Computing Machinery, Vol. 16, No. 1, January 1969, pp. 3–12.
[14] Houstis, E. N. and Rice, J. R., “Future problem solving environments for computational science,” Mathematics and Computers in Simulation, Vol. 54, No. 4-5, 2000, pp. 243 – 257.